QuantiChrom™ ATPase Assay Kit
Application
- For quantitative determination of ATPase or GTPase activity and high-throughput screen for their inhibitors.
Key Features
- High sensitivity: detection of 0.007 U/L ATPase or GTPase activity.
- Fast and convenient: single reagent, homogeneous “mix-and-measure” assay allows quantitation of enzyme activity within 30 minutes.
- Robust and amenable to HTS: detection at 620nm greatly reduces potential interference by colored compounds. Z factors of >0.7 are observed in 96-well and 384-well plates. Can be readily automated on HTS liquid handling systems.
Method
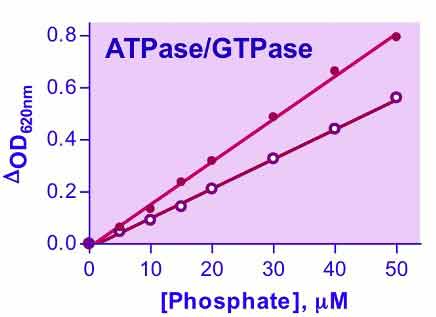
- OD620nm
Samples
- Compounds that affect ATPase/GTPase activity
Species
- All
Procedure
- 30 min
Size
- 200 tests
Detection Limit
- 0.007 U/L
Shelf Life
- 12 months
More Details
ATPases and GTPases catalyze the decomposition of ATP or GTP into ADP or GDP and free phosphate ion. These enzymes play key roles in transport, signal transduction, protein biosynthesis and cell differentiation. BioAssay Systems QuantiChrom™ ATPase/GTPase Assay Kit offers a highly sensitive method for determining ATPase/GTPase activities in a microplate format. Its proprietary formulation features a single reagent for accurate determination of enzyme activity in 30 min at room temperature. The improved malachite green reagent forms a stable dark green color with liberated phosphate, which is measured on a plate reader (600 – 660 nm).No frequently asked questions for this new product. Please check back later.
For more detailed product information and questions, please feel free to email us. Or for more general information regarding our assays, please refer to Technical Support.
De Hita, D et al. (2020). Discriminating the short-term action of root and foliar application of humic acids on plant growth: Emerging role of jasmonic acid. Frontiers in Plant Science, 11, 493. Assay: ATPase Assay Kit in cucumber roots.
Gong, D. et al. (2018). Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin. Nature communications 9(1):3623. Assay: GTPase in human hPMCA1-NPTN and hPMCA1.
Sasaki, M., Kodama, Y., Shimoyama, Y., Ishikawa, T., & Kimura, S. (2018). Aciduricity and acid tolerance mechanisms of Streptococcus anginosus. The Journal of general and applied microbiology, 2017-11. Assay: GTPase in Streptococcus anginosus NCTC 10713.
Xu, X., Ji, J., Xu, Q., Qi, X., Weng, Y., & Chen, X. (2018). The major-effect quantitative trait locus Cs ARN 6.1 encodes an AAA ATP ase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. The Plant Journal, 93(5), 917-930. Assay: GTPase in E. coli pCzn1-460 vector.
Yang, X., Chen, C., Tian, H., Chi, H., Mu, Z., Zhang, T. & Ji, X. (2018). Mechanism of ATP hydrolysis by the Zika virus helicase. The FASEB Journal, 32(10), 5250-5257. Assay: GTPase in enzymes.
Zhou, J. et al. (2018). Porcine Mx1 protein inhibits classical swine fever virus replication by targeting nonstructural protein NS5B. Journal of virology, 92(7), e02147-17. Assay: GTPase in porcine PK-15 cells.
Chen, K., Guo, L., Zhang, J., Chen, Q., Wang, K., Li, C. & Chen, D. (2017). A gene delivery system containing nuclear localization signal: Increased nucleus import and transfection efficiency with the assistance of RanGAP1. Acta biomaterialia, 48, 215-226. Assay: GTPase in mice RanGAP1.
Qian, H., Zhao, X., Cao, P., Lei, J., Yan, N., & Gong, X. (2017). Structure of the human lipid exporter ABCA1. Cell, 169(7), 1228-1239. Assay: GTPase in human ABCA1.
Sheng, Y., Song, Y., Li, Z., Wang, Y., Lin, H., Cheng, H., & Zhou, R. (2017). RAB37 interacts directly with ATG5 and promotes autophagosome formation via regulating ATG5-12-16 complex assembly. Cell Death & Differentiation 25(5):918-934. Assay: GTPase in COS-7 and HeLa cells RAB37.
Sun, S. et al. (2017). Cryo-EM structures of the ATP-bound Vps4 E233Q hexamer and its complex with Vta1 at near-atomic resolution. Nature communications, 8, 16064. Assay: GTPase in Saccharomyces cerevisiae Vps4 proteins.
Ayares, D. (2016). Multi-transgenic pigs for diabetes treatment. U.S. Patent No. 9,339,519. Assay: GTPase in porcine fetal fibroblasts.
Cao, X., Li, Y., Jin, X., Li, Y., Guo, F., & Jin, T. (2016). Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design. Nucleic acids research 44(21):10505-10514. Assay: GTPase in ZIKV NS3 helicase.
Cho, J. H., Lee, J. H., Park, Y. K., Choi, M. N., & Kim, K. N. (2016). Calcineurin B-like protein CBL10 directly interacts with TOC34 (Translocon of the Outer membrane of the Chloroplasts) and decreases its GTPase activity in Arabidopsis. Frontiers in Plant Science 7, 1911. Assay: GTPase in E. coli TOC34 protein.
El-Shesheny, I., Hijaz, F., El-Hawary, I., Mesbah, I., & Killiny, N. (2016). Impact of different temperatures on survival and energy metabolism in the Asian citrus psyllid, Diaphorina citri Kuwayama. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 192, 28-37. Assay: GTPase in Diaphorina citri hsp 70.
Killiny, N., Hijaz, F., Ebert, T. A., & Rogers, M. E. (2016). Plant bacterial pathogen manipulates the energy metabolism of its insect vector. Applied and Environmental Microbiology 83(5). pii: e03005-16. Assay: GTPase in Diaphorina citri.
Seyedmohammad, S., Fuentealba, N. A., Marriott, R. A., Goetze, T. A., Edwardson, J. M., Barrera, N. P., & Venter, H. (2016). Structural model of FeoB, the iron transporter from Pseudomonas aeruginosa, predicts a cysteine lined, GTP-gated pore. Bioscience reports, 36(2), e00322. Assay: GTPase in Pseudomonas aeruginosa FeoB Transporter.
Tian, H. et al. (2016). Structural basis of Zika virus helicase in recognizing its substrates. Protein & cell, 7(8), 562-570. Assay: GTPase in ZIKV helicase172-617.
Yang, J. et al (2016). Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule. Nature communications, 7, 12103. Assay: GTPase in E. coli tubulin.
Bertol JW, et al (2011). Antiherpes activity of glucoevatromonoside, a cardenolide isolated from a Brazilian cultivar of Digitalis lanata. Antiviral Res. 92(1):73-80. Assay: ATPase/GTPase in human, monkey Na(+)K(+)ATPase.
Binder M, et al (2011). Molecular Mechanism of Signal Perception and Integration by the Innate Immune Sensor Retinoic Acid-inducible Gene-I (RIG-I). J Biol Chem. 286(31):27278-87. Assay: ATPase/GTPase in human purified protein.
Hussey, GS et al (2011). Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 41(4):419-31. Assay: ATPase/GTPase in human GTPase.
Sheikha GA, et al (2011). Some sulfonamide drugs inhibit ATPase activity of heat shock protein 90: investigation by docking simulation and experimental validation. J Enzyme Inhib Med Chem. 26.5: 603-609. Assay: ATPase/GTPase in human Hsp90 inhibition.
Sheikha, GA et al (2010). Some sulfonamide drugs inhibit ATPase activity of heat shock protein 90: investigation by docking simulation and experimental validation. J Enzyme Inhib Med Chem. 2010 Dec 29. [Epub ahead of print]. Assay: ATPase/GTPase in human Hsp90 inhibition.
Suematsu T, et al (2010). A bacterial elongation factor G homologue exclusively functions in ribosome recycling in the spirochaete Borrelia burgdorferi. Mol Microbiol. 75(6):1445-54. Assay: ATPase/GTPase in bacteria/human ATPase.
Hu WW, et al (2009). Digoxigenin modification of adenovirus to spatially control gene delivery from chitosan surfaces. J Control Release. 135(3):250-8. Assay: ATPase/GTPase in ATPase extract.
Gong, D. et al. (2018). Structure of the human plasma membrane Ca2+-ATPase 1 in complex with its obligatory subunit neuroplastin. Nature communications 9(1):3623. Assay: GTPase in human hPMCA1-NPTN and hPMCA1.
Sasaki, M., Kodama, Y., Shimoyama, Y., Ishikawa, T., & Kimura, S. (2018). Aciduricity and acid tolerance mechanisms of Streptococcus anginosus. The Journal of general and applied microbiology, 2017-11. Assay: GTPase in Streptococcus anginosus NCTC 10713.
Xu, X., Ji, J., Xu, Q., Qi, X., Weng, Y., & Chen, X. (2018). The major-effect quantitative trait locus Cs ARN 6.1 encodes an AAA ATP ase domain-containing protein that is associated with waterlogging stress tolerance by promoting adventitious root formation. The Plant Journal, 93(5), 917-930. Assay: GTPase in E. coli pCzn1-460 vector.
Yang, X., Chen, C., Tian, H., Chi, H., Mu, Z., Zhang, T. & Ji, X. (2018). Mechanism of ATP hydrolysis by the Zika virus helicase. The FASEB Journal, 32(10), 5250-5257. Assay: GTPase in enzymes.
Zhou, J. et al. (2018). Porcine Mx1 protein inhibits classical swine fever virus replication by targeting nonstructural protein NS5B. Journal of virology, 92(7), e02147-17. Assay: GTPase in porcine PK-15 cells.
Chen, K., Guo, L., Zhang, J., Chen, Q., Wang, K., Li, C. & Chen, D. (2017). A gene delivery system containing nuclear localization signal: Increased nucleus import and transfection efficiency with the assistance of RanGAP1. Acta biomaterialia, 48, 215-226. Assay: GTPase in mice RanGAP1.
Qian, H., Zhao, X., Cao, P., Lei, J., Yan, N., & Gong, X. (2017). Structure of the human lipid exporter ABCA1. Cell, 169(7), 1228-1239. Assay: GTPase in human ABCA1.
Sheng, Y., Song, Y., Li, Z., Wang, Y., Lin, H., Cheng, H., & Zhou, R. (2017). RAB37 interacts directly with ATG5 and promotes autophagosome formation via regulating ATG5-12-16 complex assembly. Cell Death & Differentiation 25(5):918-934. Assay: GTPase in COS-7 and HeLa cells RAB37.
Sun, S. et al. (2017). Cryo-EM structures of the ATP-bound Vps4 E233Q hexamer and its complex with Vta1 at near-atomic resolution. Nature communications, 8, 16064. Assay: GTPase in Saccharomyces cerevisiae Vps4 proteins.
Ayares, D. (2016). Multi-transgenic pigs for diabetes treatment. U.S. Patent No. 9,339,519. Assay: GTPase in porcine fetal fibroblasts.
Cao, X., Li, Y., Jin, X., Li, Y., Guo, F., & Jin, T. (2016). Molecular mechanism of divalent-metal-induced activation of NS3 helicase and insights into Zika virus inhibitor design. Nucleic acids research 44(21):10505-10514. Assay: GTPase in ZIKV NS3 helicase.
Cho, J. H., Lee, J. H., Park, Y. K., Choi, M. N., & Kim, K. N. (2016). Calcineurin B-like protein CBL10 directly interacts with TOC34 (Translocon of the Outer membrane of the Chloroplasts) and decreases its GTPase activity in Arabidopsis. Frontiers in Plant Science 7, 1911. Assay: GTPase in E. coli TOC34 protein.
El-Shesheny, I., Hijaz, F., El-Hawary, I., Mesbah, I., & Killiny, N. (2016). Impact of different temperatures on survival and energy metabolism in the Asian citrus psyllid, Diaphorina citri Kuwayama. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology 192, 28-37. Assay: GTPase in Diaphorina citri hsp 70.
Killiny, N., Hijaz, F., Ebert, T. A., & Rogers, M. E. (2016). Plant bacterial pathogen manipulates the energy metabolism of its insect vector. Applied and Environmental Microbiology 83(5). pii: e03005-16. Assay: GTPase in Diaphorina citri.
Seyedmohammad, S., Fuentealba, N. A., Marriott, R. A., Goetze, T. A., Edwardson, J. M., Barrera, N. P., & Venter, H. (2016). Structural model of FeoB, the iron transporter from Pseudomonas aeruginosa, predicts a cysteine lined, GTP-gated pore. Bioscience reports, 36(2), e00322. Assay: GTPase in Pseudomonas aeruginosa FeoB Transporter.
Tian, H. et al. (2016). Structural basis of Zika virus helicase in recognizing its substrates. Protein & cell, 7(8), 562-570. Assay: GTPase in ZIKV helicase172-617.
Yang, J. et al (2016). Pironetin reacts covalently with cysteine-316 of alpha-tubulin to destabilize microtubule. Nature communications, 7, 12103. Assay: GTPase in E. coli tubulin.
Bertol JW, et al (2011). Antiherpes activity of glucoevatromonoside, a cardenolide isolated from a Brazilian cultivar of Digitalis lanata. Antiviral Res. 92(1):73-80. Assay: ATPase/GTPase in human, monkey Na(+)K(+)ATPase.
Binder M, et al (2011). Molecular Mechanism of Signal Perception and Integration by the Innate Immune Sensor Retinoic Acid-inducible Gene-I (RIG-I). J Biol Chem. 286(31):27278-87. Assay: ATPase/GTPase in human purified protein.
Hussey, GS et al (2011). Identification of an mRNP complex regulating tumorigenesis at the translational elongation step. Mol Cell. 41(4):419-31. Assay: ATPase/GTPase in human GTPase.
Sheikha GA, et al (2011). Some sulfonamide drugs inhibit ATPase activity of heat shock protein 90: investigation by docking simulation and experimental validation. J Enzyme Inhib Med Chem. 26.5: 603-609. Assay: ATPase/GTPase in human Hsp90 inhibition.
Sheikha, GA et al (2010). Some sulfonamide drugs inhibit ATPase activity of heat shock protein 90: investigation by docking simulation and experimental validation. J Enzyme Inhib Med Chem. 2010 Dec 29. [Epub ahead of print]. Assay: ATPase/GTPase in human Hsp90 inhibition.
Suematsu T, et al (2010). A bacterial elongation factor G homologue exclusively functions in ribosome recycling in the spirochaete Borrelia burgdorferi. Mol Microbiol. 75(6):1445-54. Assay: ATPase/GTPase in bacteria/human ATPase.
Hu WW, et al (2009). Digoxigenin modification of adenovirus to spatially control gene delivery from chitosan surfaces. J Control Release. 135(3):250-8. Assay: ATPase/GTPase in ATPase extract.
To find more recent publications, please click here.
If you or your labs do not have the equipment or scientists necessary to run this assay, BioAssay Systems can perform the service for you.
– Fast turnaround
– Quality data
– Low cost
Please email or call 1-510-782-9988 x 2 to discuss your projects.
$469.00
For bulk quote or custom reagents, please email or call 1-510-782-9988 x 1.
Orders are shipped the same day if placed by 2pm PST
Shipping: RT
Carrier: Fedex
Delivery: 1-2 days (US), 3-6 days (Intl)
Storage: 4°C upon receipt
Related Products
You may also like…
| Name | SKU | Price | Buy |
|---|---|---|---|
| QuantiChrom™ Acetylcholinesterase Inhibitor Screening Kit | IACE-100 |
$449.00 |
Agriculture & Environment
QuantiChrom™ Acetylcholinesterase Inhibitor Screening Kit$449.00 Add to cart |
| QuantiChrom™ Arginase Inhibitor Screening Kit | IARG-100 |
$309.00 |
|
| EnzyChrom™ Monoamine Oxidase Inhibitor Screening Kit | EIMO-100 |
$459.00 |
|
| EnzyChrom™ Nitric Oxide Synthase Inhibitor Screening Kit | EINO-100 |
$529.00 |
Why BioAssay Systems
Quality and User-friendly • Expert Technical Support • Competitive Prices • Expansive Catalogue • Trusted Globally
